Written by: Sheng Ming
Translated by: Li Zebing
Edited by: William Mosteller
Date: 1-26
Harbin Institute of Technology News (Sheng Ming/Text) Recently, Professor Huang Zhiwei's research group in the School of Life and Technology of Harbin Institute of Technology revealed the molecular mechanism of the compatibility and high fidelity of a SpCas9 variant (xCas9 3.7) in recognizing substrates through structural biology and biochemical research. The study was published in the journal Cell Research under the title "Structural insights into a high fidelity variant of SpCas9".
The CRISPR-Cas system is a bacterial-encoded adaptive immune system that defends against bacteriophages. The system cleaves viral DNA or RNA by RNA-guided effector proteins to resist bacteriophage (viral) infection. The combination of Streptococcus pyogenes (SpCas9) and sgRNA has the ability to efficiently and conveniently "edit" the target gene in cells or organisms, so the CRISPR-Cas9 system and its derivatives have been rapidly applied to gene editing in recent years. Although the CRISPR-Cas9 system is widely used as a gene-editing tool in biology, medicine, and agriculture, the miss-targeting effect and the restriction on the recognition of PAM by the CRISPR-Cas9 system is still an urgent problem to be solved. At present, high fidelity SpCas9 mutants such as xCas9 3.7 (A262T, R324L, S409I, E480K, E543D, M694I, E1219V) have been produced by screening for SpCas9 protein. However, the high fidelity structural basis of SpCas9 mutants is unknown.
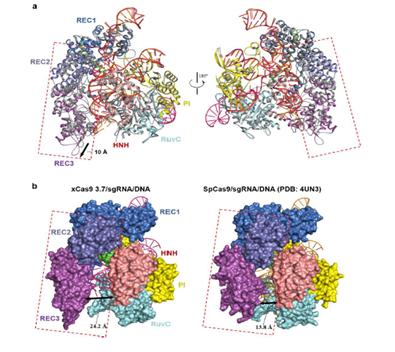
In order to reveal the molecular mechanism of xCas93.7 with extensive PAM compatibility and high DNA targeting specificity, the team analyzed the three-dimensional structure of xCas9 3.7, sgRNA, and the ternary complex containing two different PAM double-stranded DNA. It was found that E1219 was critical for SpCas9 to select the PAM sequence strictly through salt bridge stabilized R1335. The mutation of E1219V in xCas9 3.7 lifted the restriction on the side chain of R1335, giving it a certain degree of freedom, and thus increasing the PAM recognition ability in other substrates.
Compared with wild-type (WT) SpCas9, xCas9 3.7 has significant conformational changes in the REC2 and REC3 domains, although the overall structure of the two is similar, which reduces the contact with DNA substrates and increases the specificity of substrate recognition, thus reducing the miss-targeting effect of xCas9 3.7. Based on the mechanism found in this study, the team rationally designed a series of variants of SpCas9, which showed significant improvements in fidelity in biochemical and cellular experiments compared with wild-type SpCas9 mutants. This study not only reveals the molecular mechanism of a high fidelity Cas9 gene editing system for the first time, but also provides a key structural basis for modifying SpCas9 and other Cas endonucleases to become more efficient and specific gene editing tools, which has the application value of guiding the modification of new gene editing systems. This is another achievement of the team in the field of pathogen-host interaction systems and gene editing systems.
Professor Huang Zhiwei is the correspondent author of the paper. Dr Guo Minghui, Ren Kuan, and postdoctoral teacher Zhu Yuwei are the co-authors of the paper. Shanghai Synchrotron Radiation Center provided support for crystal data collection. The program is supported by the National Natural Science Foundation of China, Harbin Institute of Technology Young Scientist Studio, and other funds.
Link to article: https://www.nature.com/articles/s41422-018-0131-6