Written by: Sheng Ming
Translated by: Li Zebing
Edited by: William Mosteller
Date: 2019-4-2
Harbin Institute of Technology News (Sheng Ming/Text) On April 1, Professor Huang Zhiwei of the School of Life and Technology of Harbin Institute of Technology discovered a new type of anti-CRISPR protein and revealed its new mechanism of inhibiting type V cas12a activity. The work was published in the journal Nature Structural & Molecular Biology under the title "An anti-CRISPR protein types V Cas12a by acetylation", and was featured in the News & Views column of the journal.
The CRISPR-Cas adaptive immune system provides nucleic acid sequence-specific defense mechanisms against bacteriophage and plasmid invasion by bacteria and archaea. The CRISPR-Cas system can be divided into six subtypes. Type II Cas9 and V Cas12a (Cpf1) systems are widely used in genome editing and various biotechnology applications. In the process of bacteriophage infection, a bacterial Cas effector protein (Cas9 or Cas12a) recognizes the PAM sequence of targeting double-stranded DNA through PAM-interacting (PI) domain under the guidance of RNA and unscrews the two strands of targeting dsDNA, thus cleaving the two strands of targeting dsDNA and resisting the invasion of bacteriophage. As a counterattack to the CRISPR-Cas immune system, bacteriophages have evolved anti-CRISPR proteins to escape from the bacterial CRISPR-Cas defense system.
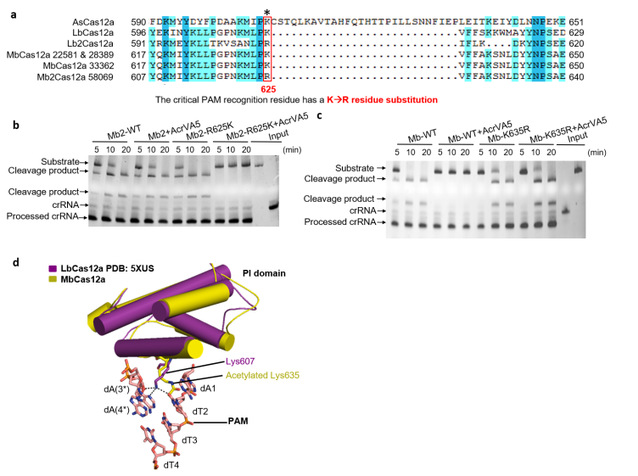
Firstly, a class of anti-Cas12a candidate proteins were identified by bioinformatics. Then, one of the anti-Cas12a proteins, AcrVA5, was found to inhibit the activity of Moraxella bovoculi (Mb) Cas12a by biochemical and bacterial experiments. Unexpectedly, the AcrVA5 protein did not bind to the MbCas12a-crRNA in a gel migration assay, which was completely different from previous studies which found that anti-CRISPR proteins inhibited the binding of targeted DNA by binding to the Cas protein or inhibiting the endonuclease activity of the Cas protein. They speculated that the inhibitory activity of MbCas12a might be mediated by an unknown enzyme modification. The AcrVA5 protein was found to have acetyltransferase activity by sequence analysis and experiments. Further biochemical experiments confirmed that the AcrVA5 protein inactivates the MbCas12a protein through acetyltransferase activity.
The team then searched for the acetylation sites of AcrVA5 on the MbCas12a protein. These acetyltransferases usually modify lysine (K) residues on substrate proteins, and since K635 is the key amino acid in the PAM sequence of MbCas12a's PI domain recognizing the substrate DNA, they speculated that K635 of MbCas12a might be the key site for the acetylation modification of AcrVA5 to inactivate it. The K635R mutation assay of MbCas12a confirmed that the mutant activity could not be inhibited by AcrVA5. It was also found that AcrVA5 had no inhibitory effect on the Cas12a of another Mb strain (Mb2) (arginine, not lysine, was the key amino acid for PAM recognition). These results indicated that the inhibitory effect of the AcrVA5 protein was mainly dependent on the K residue of the PAM recognition site of the acetylated Cas12a protein. Interestingly, the activity of the R625K mutant of Mb2Cas12a was completely inhibited by AcrVA5, which combined with mass spectrometry analysis, confirmed that the AcrVA5 protein was inactivated by modifying the key K635 residue of PAM, the recognition substrate of the MbCas12a protein. The structure of the Cas12a protein revealed that the acetylation of K635 not only caused the loss of hydrogen bond interaction between dT2 and dA3*, but also resulted in the formation of steric hindrance, which prevented the binding of dsDNA, thus depriving Cas12a of dsDNA cleaving activity.
The mechanism of anti-CRISPR modifying and inhibiting CRISPR protein by acetyltransferase activity revealed in this study is completely different from that of anti-Cas9, which inhibits Cas9 activity by directly binding to the substrate binding sites of Cas9. This study reveals for the first time that anti-CRISPR protein can inhibit the immune defense system of bacterial CRISPR-Cas through enzyme modification, and provides a new perspective for the attack defense strategy between bacteriophage and bacterial immune system (CRISPR-Cas). This discovery can be used to precisely control the activity of Cas12a in vivo and improve the accuracy of gene editing. This is another important achievement of the team in the field of pathogen-host interaction and the molecular mechanism of gene editing.
Professor Huang Zhiwei is the correspondent author of the research paper, while doctoral candidate Dong Liyonghe and Guan Xiaoyu, and doctoral Li Ningning from the Gao Ning group of Peking University are the co-authors of the paper. Dr. Zhu Yuwei and associate professor Zhang Fan of Harbin Institute of Technology made an important contribution to the study. The Shanghai Synchrotron Radiation Center provides support for crystal data collection. The program is supported by the National Natural Science Foundation of China, Harbin Institute of Technology Young Scientist Studio, and other funds.
Link to article:https://www.nature.com/articles/s41594-019-0206-1