On June 9th, the research group of Prof. Huang Zhiwei from School of Life Science and Technology of HIT published a research article entitled “Structural basis of Staphylococcus aureus Cas9 inhibition by AcrIIA14” online in Nucleic Acids Research. In this study, the high-resolution crystal structure of the Anti-CRISPR protein, namely complex of AcrIIA14 bound SauCas9 (SauCas9) was analyzed. Through structural analysis and corresponding biochemical experiments, the molecular basis of specifical inhibition of SauCas9 cleavage by AcrIIA14 was explained. This study widens the understanding of regulation of CRISPR-CAS system by Anti-CRISPR protein, and provides theoretical basis and new ideas for accurately controlling SauCas9 and applications of other CRISPR-CAS systems for gene editing.
CRISPR-CAS system has become the most widely used gene editing tool due to its convenient operability and efficient in vivo editing. However, there are still many problems that need to be solved urgently in practical application, one of which is the miss effect of gene editing due to its uncontrollable clevage activity. How to put the brakes on the gene editing system and control its editing activity has always been a problem that the field is trying to solve. As an adaptive immune system of bacteria, CRISPR-Cas system is used for phage invasion. Facing the selection pressure of bacterial CRISPR-Cas, phage evolved a corresponding antagonistic mechanism, in which case the phage evolved Anti-CRISPR protein to inhibit the activity of bacterial CRISPR-Cas system. In previous studies, it was found that Anti-CRISPR protein AcrIIA14 can inhibit the activity of SauCas9, but the specific molecular basis is not clear.
In this study, a stable quaternary complex protein of AcrIIA14 combined with SauCas9-sgRNA-dsDNA was purified and assembled in vitro, and then the crystal structure of the complex was analyzed by X-ray crystallography. By analyzing the structure of the complex, it was found that AcrIIA14 protein interacted with HNH domain of SauCas9 protein in a stoichiometric ratio of 1: 1. AcrIIA14 protein binds to HNH nuclease domain, which makes it undergo conformational change. This conformational change of HNH causes its active site to be far away from the binding target DNA, thus inhibiting the activation of SauCas9. In addition, through structural comparison, it was also found that the binding of AcrIIA14 induced the allosterism of HNH domain and L1 linker in SauCas9, resulting in two new intramolecular interaction interfaces. Combined with biochemical experiments, it was found that the allosterism of SauCas9 induced by AcrIIA14 enhanced the inhibitory effect. This effect of inducing Cas9 protein allosterism is different from the other two inhibitors AcrIIC1 and AcrIIC3 found in the past, which also bind to the HNH domain, showing the diversity of anti-CRISPR protein inhibiting SauCas9.
Liu Hongnan, a Ph.D. student at the School of Life Science and Technology, and Zhu Yuwei, an associate research fellow at the School of Life Science and Technology, are the first authors of the paper; Prof. Huang Zhiwei is the correspondence author of this research paper; Lu Zebin, a graduate student, participated in part of the research. The crystal diffraction data of this study were collected at Shanghai Light Source BL17U. This project is supported by the National Natural Science Foundation of China.
Link of the Article:https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab487/6295537
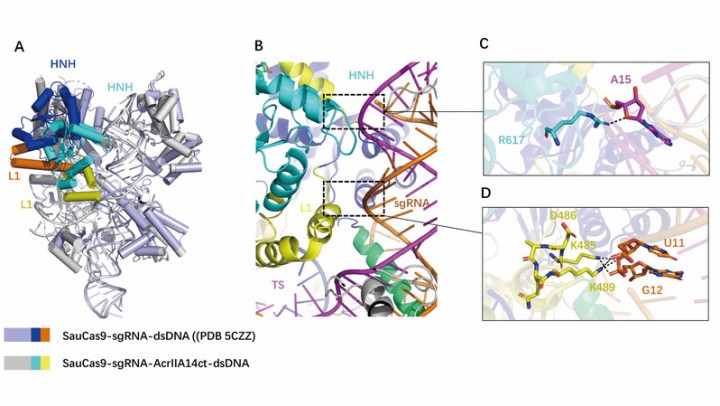
AcrIIA14ct binding induces SauCas9 allostery


